Embedding of nodes happens via word2vec by means of a smart trick: using randomg walks over the graph to generate ‘word’ sequences.
Stellargraph has its own direct method to perform the embedding but the intermediate methods highlights better the process. So, below we generate the node2vec embedding via an explicit walk and show how it generates a really good community detection separation.
import networkx as nx
import numpy as np
import matplotlib.pyplot as plt
from sklearn.manifold import TSNE
%matplotlib inline
We’ll use the karater club to demonstrate the process. The graph consists of two sets of nodes which are a well-separated according to the ‘club’ property.
g_nx = nx.karate_club_graph()
cols = ["green" if g_nx.nodes[n]["club"]=='Officer' else "orange" for n in g_nx.nodes()]
nx.draw(g_nx, node_color=cols)
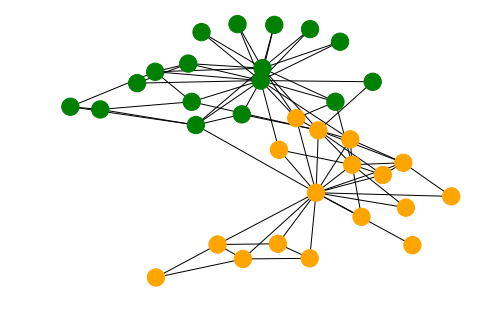
From this graph we create a Stellargraph and perform a biased random walk on it. This generates word sequences, in this case the string value of the node index.
from stellargraph.data import BiasedRandomWalk
from stellargraph import StellarGraph
from gensim.models import Word2Vec
rw = BiasedRandomWalk(StellarGraph(g_nx))
walks = rw.run(
nodes=list(g_nx.nodes()), # root nodes
length=100, # maximum length of a random walk
n=10, # number of random walks per root node
p=0.5, # Defines (unormalised) probability, 1/p, of returning to source node
q=2.0 # Defines (unormalised) probability, 1/q, for moving away from source node
)
walks = [list(map(str, walk)) for walk in walks]
model = Word2Vec(walks, size=128, window=5, min_count=0, sg=1, workers=2, iter=1)
The value of an embedding is for instance
model.wv['29']
array([ 0.0283457 , 0.06906749, -0.09740856, 0.08761664, 0.0240158 ,
-0.04252268, 0.05366189, 0.12255755, -0.14192946, -0.12441556,
0.14022443, 0.16821992, 0.01899681, 0.02525605, -0.129657 ,
-0.00075872, -0.10963597, -0.24603637, 0.14481993, 0.04069758,
...
-0.03686432, 0.28888953, 0.06754036], dtype=float32)
In order to visualize the embedding one has to somehow reduce the dimension. This is most easily done via t-SNE.
# Retrieve node embeddings and corresponding subjects
node_ids = model.wv.index2word # list of node IDs
node_embeddings = model.wv.vectors # numpy.ndarray of size number of nodes times embeddings dimensionality
node_targets = [ g_nx.node[int(node_id)]['club'] for node_id in node_ids]
# Apply t-SNE transformation on node embeddings
tsne = TSNE(n_components=2)
node_embeddings_2d = tsne.fit_transform(node_embeddings)
alpha=0.9
label_map = { l: i for i, l in enumerate(np.unique(node_targets))}
node_colours = [ label_map[target] for target in node_targets]
plt.figure(figsize=(10,8))
plt.scatter(node_embeddings_2d[:,0],
node_embeddings_2d[:,1],
c=node_colours, cmap="jet", alpha=alpha)
<matplotlib.collections.PathCollection at 0x1463cfc50>
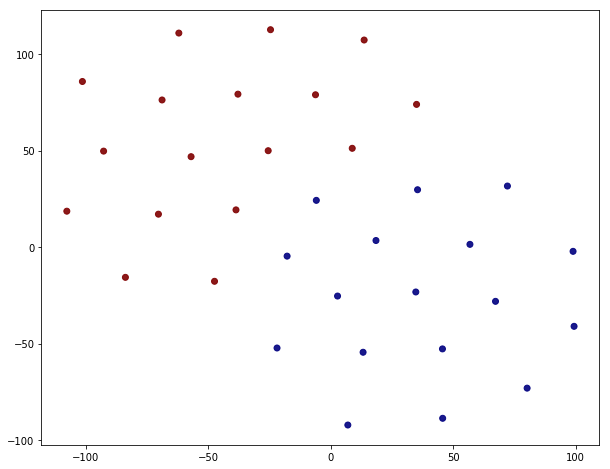
This looks like a clean separation indeed. The splitting is not 100% correct though, just by looking at the corresponding value of the ‘club’ property.
[g_nx.nodes[i] for i,v in enumerate(node_colours) if v==1]
[{'club': 'Mr. Hi'},
{'club': 'Mr. Hi'},
{'club': 'Mr. Hi'},
{'club': 'Officer'},
{'club': 'Mr. Hi'},
{'club': 'Officer'},
{'club': 'Officer'},
{'club': 'Officer'},
{'club': 'Mr. Hi'},
{'club': 'Officer'},
{'club': 'Officer'},
{'club': 'Officer'},
{'club': 'Officer'},
{'club': 'Officer'},
{'club': 'Officer'},
{'club': 'Officer'},
{'club': 'Officer'}]
Five out of senteen are incorrect. This is still remarkable considering the fact that node2vec process did not know anything at about the ‘club’ property but that it’s an emergent feature of the embedding.

